Mouse P-Cadherin Antibody Summary
Glu100-Gly647
Accession # Q8BSL6
*Small pack size (-SP) is supplied either lyophilized or as a 0.2 µm filtered solution in PBS.
Applications
Mouse P-Cadherin Sandwich Immunoassay
Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website.
Scientific Data
 View Larger
View Larger
Detection of P‑Cadherin by Western Blot. Western blot shows lysates of mouse embryo tissue. PVDF membrane was probed with 0.5 µg/mL of Goat Anti-Mouse P-Cadherin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF761) followed by HRP-conjugated Anti-Goat IgG Secondary Antibody (HAF017). A specific band was detected for P-Cadherin at approximately 115 kDa (as indicated). This experiment was conducted under reducing conditions and using Immunoblot Buffer Group 1.
 View Larger
View Larger
P‑Cadherin in A431 Human Cell Line. P-Cadherin was detected in immersion fixed A431 human epithelial carcinoma cell line using Goat Anti-Mouse P-Cadherin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF761) at 10 µg/mL for 3 hours at room temperature. Cells were stained using the NorthernLights™ 493-conjugated Anti-Goat IgG Secondary Antibody (green; NL003) and counterstained with DAPI (blue). Specific staining was localized to intercellular junctions. View our protocol for Fluorescent ICC Staining of Cells on Coverslips.
 View Larger
View Larger
P‑Cadherin in Mouse Embryo. P-Cadherin was detected in immersion fixed frozen sections of mouse embryo (15 dpc) using Goat Anti-Mouse P-Cadherin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF761) at 15 µg/mL overnight at 4 °C. Tissue was stained using the Anti-Goat HRP-DAB Cell & Tissue Staining Kit (brown; CTS008) and counterstained with hematoxylin (blue). Specific staining was localized to connective tissue and lungs. View our protocol for Chromogenic IHC Staining of Frozen Tissue Sections.
 View Larger
View Larger
Detection of P‑Cadherin in Human Liver. P‑Cadherin was detected in immersion fixed paraffin-embedded sections of human liver using Goat Anti-Mouse P‑Cadherin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF761) at 0.5 µg/ml for 1 hour at room temperature followed by incubation with the Anti-Goat IgG VisUCyte™ HRP Polymer Antibody (Catalog # VC004). Before incubation with the primary antibody, tissue was subjected to heat-induced epitope retrieval using VisUCyte Antigen Retrieval Reagent-Basic (Catalog # VCTS021). Tissue was stained using DAB (brown) and counterstained with hematoxylin (blue). Specific staining was localized to the cell membrane of hepatocytes. View our protocol for IHC Staining with VisUCyte HRP Polymer Detection Reagents.
 View Larger
View Larger
Detection of Mouse P‑Cadherin by Simple WesternTM. Simple Western lane view shows lysates of mouse embryo tissue, loaded at 0.2 mg/mL. A specific band was detected for P-Cadherin at approximately 115 kDa (as indicated) using 25 µg/mL of Goat Anti-Mouse P-Cadherin Antigen Affinity-purified Polyclonal Antibody (Catalog # AF761) followed by 1:50 dilution of HRP-conjugated Anti-Goat IgG Secondary Antibody (HAF109). This experiment was conducted under reducing conditions and using the 12-230 kDa separation system.
 View Larger
View Larger
Detection of Mouse P-Cadherin by Western Blot BMPR1a regulated P-cadherin expression via p63 and Slug. (A) Immunohistochemistry staining for P-cadherin in control (n = 4 mice) and cKO (n = 3 mice) mammary glands at pregnancy day 14.5. Scale bar, 50 μm. (B) Western blotting for P-cadherin in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 mice. (C) qRT-PCR analysis of Cdh3 in FACS-sorted control and cKO myoepithelial cells at pregnancy day 14.5. n = 4 biological replicates. (D) P-cadherin (red) and K14 (green) double immunofluorescence staining in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. n = 3 biological replicates. Scale bar, 25 μm. (E) Western blotting for P-cadherin in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 biological replicates. (F) Scatter plot showing the correlation between BMPR1A and CDH3 expression in mammary glands from TCGA and GTEx data. Pearson’s coefficient test was performed to assess statistical significance. (G) Western blotting for P-cadherin in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Actin. n = 3 biological replicates. (H) Immunofluorescence for P-cadherin (green) in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. n = 3 biological replicates. Scale bar, 25 μm. (I) Western blotting for K14, P-cadherin, p63 and Slug in HC11 cells treated with scramble RNA, p63 siRNA, Slug siRNA, or p63 siRNA and Slug siRNA at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of K14/ beta -Actin, P-cadherin/ beta -Actin, p63/ beta -Actin and Slug/ beta -Actin. n = 3 biological replicates. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34336839), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of Mouse P-Cadherin by Western Blot BMPR1a regulated P-cadherin expression via p63 and Slug. (A) Immunohistochemistry staining for P-cadherin in control (n = 4 mice) and cKO (n = 3 mice) mammary glands at pregnancy day 14.5. Scale bar, 50 μm. (B) Western blotting for P-cadherin in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 mice. (C) qRT-PCR analysis of Cdh3 in FACS-sorted control and cKO myoepithelial cells at pregnancy day 14.5. n = 4 biological replicates. (D) P-cadherin (red) and K14 (green) double immunofluorescence staining in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. n = 3 biological replicates. Scale bar, 25 μm. (E) Western blotting for P-cadherin in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 biological replicates. (F) Scatter plot showing the correlation between BMPR1A and CDH3 expression in mammary glands from TCGA and GTEx data. Pearson’s coefficient test was performed to assess statistical significance. (G) Western blotting for P-cadherin in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Actin. n = 3 biological replicates. (H) Immunofluorescence for P-cadherin (green) in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. n = 3 biological replicates. Scale bar, 25 μm. (I) Western blotting for K14, P-cadherin, p63 and Slug in HC11 cells treated with scramble RNA, p63 siRNA, Slug siRNA, or p63 siRNA and Slug siRNA at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of K14/ beta -Actin, P-cadherin/ beta -Actin, p63/ beta -Actin and Slug/ beta -Actin. n = 3 biological replicates. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34336839), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of Mouse P-Cadherin by Western Blot BMPR1a regulated P-cadherin expression via p63 and Slug. (A) Immunohistochemistry staining for P-cadherin in control (n = 4 mice) and cKO (n = 3 mice) mammary glands at pregnancy day 14.5. Scale bar, 50 μm. (B) Western blotting for P-cadherin in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 mice. (C) qRT-PCR analysis of Cdh3 in FACS-sorted control and cKO myoepithelial cells at pregnancy day 14.5. n = 4 biological replicates. (D) P-cadherin (red) and K14 (green) double immunofluorescence staining in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. n = 3 biological replicates. Scale bar, 25 μm. (E) Western blotting for P-cadherin in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 biological replicates. (F) Scatter plot showing the correlation between BMPR1A and CDH3 expression in mammary glands from TCGA and GTEx data. Pearson’s coefficient test was performed to assess statistical significance. (G) Western blotting for P-cadherin in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Actin. n = 3 biological replicates. (H) Immunofluorescence for P-cadherin (green) in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. n = 3 biological replicates. Scale bar, 25 μm. (I) Western blotting for K14, P-cadherin, p63 and Slug in HC11 cells treated with scramble RNA, p63 siRNA, Slug siRNA, or p63 siRNA and Slug siRNA at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of K14/ beta -Actin, P-cadherin/ beta -Actin, p63/ beta -Actin and Slug/ beta -Actin. n = 3 biological replicates. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34336839), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of Mouse P-Cadherin by Western Blot BMPR1a regulated P-cadherin expression via p63 and Slug. (A) Immunohistochemistry staining for P-cadherin in control (n = 4 mice) and cKO (n = 3 mice) mammary glands at pregnancy day 14.5. Scale bar, 50 μm. (B) Western blotting for P-cadherin in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 mice. (C) qRT-PCR analysis of Cdh3 in FACS-sorted control and cKO myoepithelial cells at pregnancy day 14.5. n = 4 biological replicates. (D) P-cadherin (red) and K14 (green) double immunofluorescence staining in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. n = 3 biological replicates. Scale bar, 25 μm. (E) Western blotting for P-cadherin in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 biological replicates. (F) Scatter plot showing the correlation between BMPR1A and CDH3 expression in mammary glands from TCGA and GTEx data. Pearson’s coefficient test was performed to assess statistical significance. (G) Western blotting for P-cadherin in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Actin. n = 3 biological replicates. (H) Immunofluorescence for P-cadherin (green) in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. n = 3 biological replicates. Scale bar, 25 μm. (I) Western blotting for K14, P-cadherin, p63 and Slug in HC11 cells treated with scramble RNA, p63 siRNA, Slug siRNA, or p63 siRNA and Slug siRNA at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of K14/ beta -Actin, P-cadherin/ beta -Actin, p63/ beta -Actin and Slug/ beta -Actin. n = 3 biological replicates. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34336839), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of Mouse P-Cadherin by Western Blot BMPR1a regulated P-cadherin expression via p63 and Slug. (A) Immunohistochemistry staining for P-cadherin in control (n = 4 mice) and cKO (n = 3 mice) mammary glands at pregnancy day 14.5. Scale bar, 50 μm. (B) Western blotting for P-cadherin in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 mice. (C) qRT-PCR analysis of Cdh3 in FACS-sorted control and cKO myoepithelial cells at pregnancy day 14.5. n = 4 biological replicates. (D) P-cadherin (red) and K14 (green) double immunofluorescence staining in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. n = 3 biological replicates. Scale bar, 25 μm. (E) Western blotting for P-cadherin in HC11 mammary epithelial cells treated with BMP4 (50 ng/mL) for 24 h. beta -Tubulin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Tubulin. n = 3 biological replicates. (F) Scatter plot showing the correlation between BMPR1A and CDH3 expression in mammary glands from TCGA and GTEx data. Pearson’s coefficient test was performed to assess statistical significance. (G) Western blotting for P-cadherin in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of P-cadherin/ beta -Actin. n = 3 biological replicates. (H) Immunofluorescence for P-cadherin (green) in HC11 cells treated with p63 siRNA (sip63)/Slug siRNA (siSlug) and scramble RNA (NC) at 48 h. n = 3 biological replicates. Scale bar, 25 μm. (I) Western blotting for K14, P-cadherin, p63 and Slug in HC11 cells treated with scramble RNA, p63 siRNA, Slug siRNA, or p63 siRNA and Slug siRNA at 48 h. beta -Actin was used as a loading control. Statistical analysis the expression of K14/ beta -Actin, P-cadherin/ beta -Actin, p63/ beta -Actin and Slug/ beta -Actin. n = 3 biological replicates. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/34336839), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
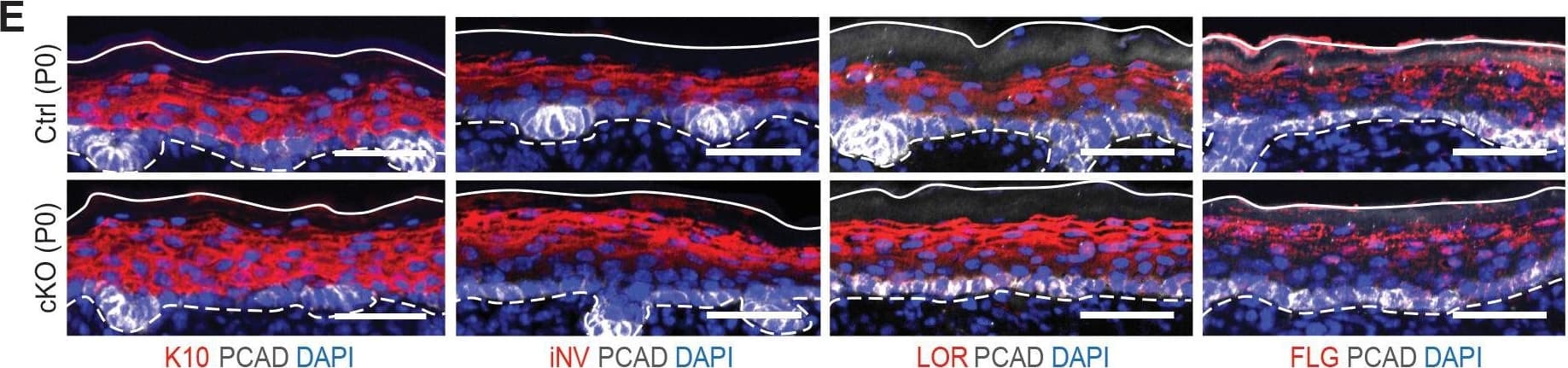
Detection of Mouse P-Cadherin by Immunohistochemistry Additional analysis of epidermal perturbations upon Mettl3 cKO. (E) Images of P0 back skin sagittal sections immulabeled for K10, involucrin (iNV), loricrin (LOR)&filaggrin (FLG) demonstrating the expression pattern of standard differentiation markers is not severely interrupted by Mettl3 cKO (scale bars: 50 µm). Solid&dashed lines as in (B). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
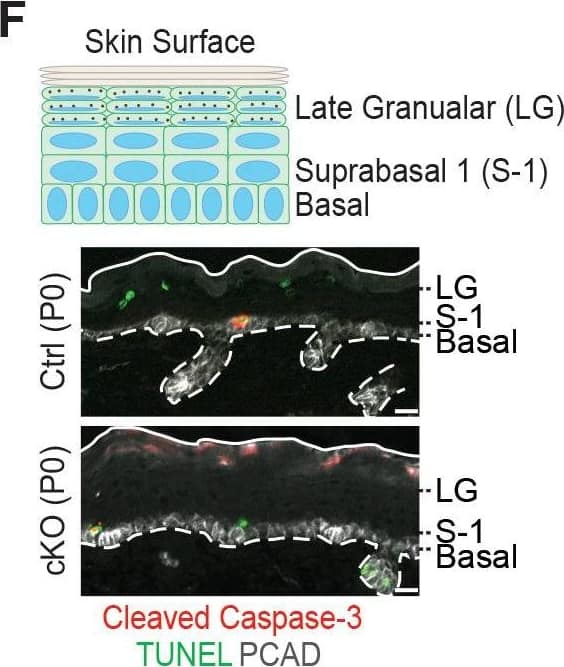
Detection of Mouse P-Cadherin by Immunohistochemistry Additional analysis of epidermal perturbations upon Mettl3 cKO. (F) Representative images of P0 back skin sagittal sections with cleaved Caspase-3 immunostaining&TUNEL staining (scale bars: 20 µm). White solid lines indicate skin surface&dashed lines indicate dermal-epithelial border. Quantification of TUNEL+&cleaved Caspase-3+ cells in the designated layers at P0. Only double-positive cells denote apoptosis; TUNEL+ only cells are typically seen as granular cells lose their nuclei&transition as dead flattened cells to the stratum corneum. This is defective in Mettl3 cKO skin. The TUNEL+ cells in the suprabasal 1 layer likely correspond to the occasional cytolytic suprabasal cells that we saw at the ultrastructural level (not shown). Quantification of the labeled cells is shown (error bars: standard deviation, for each condition n = 3 biological replicates × 10 images per replicate, *p<0.05 and **p<0.01 by unpaired two-tailed Student’s t-test).Figure 6—figure supplement 2—source data 1.Quantification of PCAD, ECAD immunofluorescence signals in (A).Figure 6—figure supplement 2—source data 2.Quantification of EdU+ cells&the suprabasal/basal cell number ratio in (B).Figure 6—figure supplement 2—source data 3.Quantification of cell sizes by cytospin in (C).Figure 6—figure supplement 2—source data 4.Quantification of cell death events in epidermis in (F).Quantification of PCAD, ECAD immunofluorescence signals in (A).Quantification of EdU+ cells&the suprabasal/basal cell number ratio in (B).Quantification of cell sizes by cytospin in (C).Quantification of cell death events in epidermis in (F). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
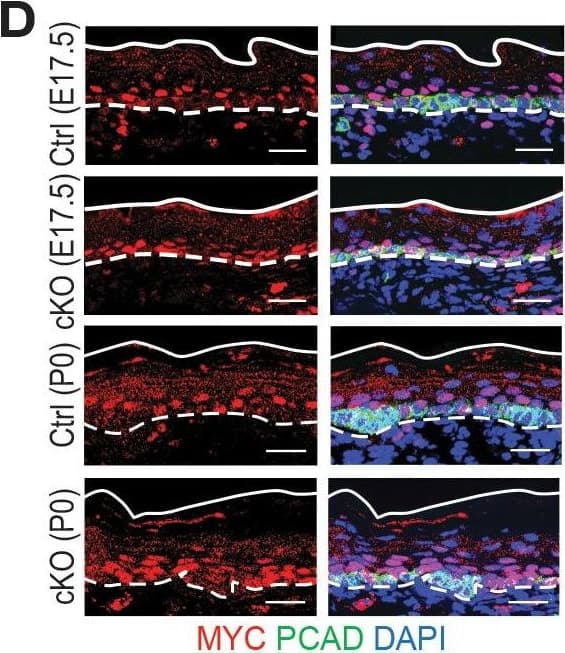
Detection of Mouse P-Cadherin by Immunohistochemistry Additional analysis of features affected by the upregulated genes upon Mettl3 cKO.(A) ECDF plots of the Z score (cKO/Ctrl) from scRNA-seq demonstrate transcripts with higher levels of m6A modification (assessed by the coding sequence SN-uTPM per nt value from miCLIP) in wild-type tend to be more upregulated upon METTL3 loss. The p values were calculated through Wilcoxon rank sum test. (B, C) Violin plots illustrating the relative expression levels of Bmyc and Myc mRNA in corresponding groups of Mettl3 cKO versus control cells. Z score assessment of expressional difference between cKO and control [Z (cKO/Ctrl)] and false discovery rate (FDR) is calculated by MAST. The upregulation is verified with qPCR on total RNA samples extracted from YFP+ skin epithelial cells FACS isolated from E16.5 embryos with Tbp mRNA as internal control (error bars: standard deviation, for each condition n = 3 biological replicates, **p<0.01 by unpaired two-tailed Student’s t-test). (D) Images from E17.5 and P0 sagittal sections immunolabeled for MYC and PCAD (scale bars: 25 µm). White solid lines denote skin surface and dashed lines denote dermal:epidermal border. Quantifications of MYC immunofluorescence specifically in the Epi basal cells (identified based on PCAD staining) reveals the upregulation of MYC in cKO samples (for each condition n = 3 biological replicates ×7 images per replicate, *p<0.05 and ***p<0.001 by unpaired two-tailed Student’s t-test).Figure 7—figure supplement 1—source data 1.Bmyc qPCR in (B).Figure 7—figure supplement 1—source data 2.Myc qPCR in (C).Figure 7—figure supplement 1—source data 3.Quantification of MYC immunofluorescence signals in (D).Bmyc qPCR in (B).Myc qPCR in (C).Quantification of MYC immunofluorescence signals in (D). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
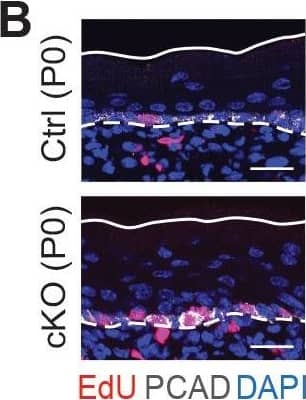
Detection of Mouse P-Cadherin by Immunohistochemistry Additional analysis of epidermal perturbations upon Mettl3 cKO. (B) Left panel: representative images from P0 sagittal sections with a 45-min EdU pulse prior to tissue collection (scale bars: 25 µm). Solid lines, skin surface boundary; dashed lines, dermal-epidermal border. Middle panel: quantification of the ratio of EdU+ cells among all basal cells (for each condition n = 3 biological replicates ×10 images per replicate, ***p<0.001 by unpaired two-tailed Student’s t-test). Right panel: quantification of the suprabasal cell number/basal cell number ratio according to DAPI staining of the nuclei (for each condition n = 5 biological replicates ×10 images per replicate, ***p<0.001 by unpaired two-tailed Student’s t-test). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
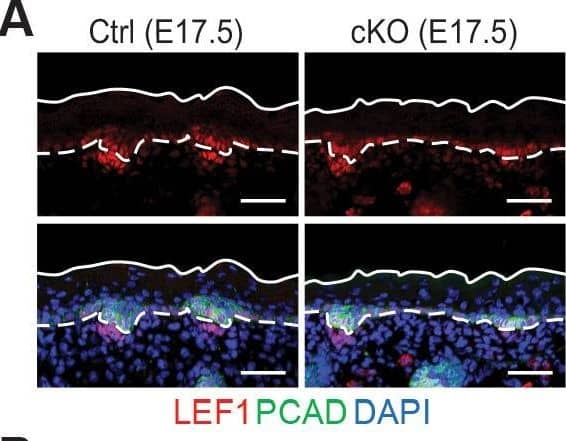
Detection of Mouse P-Cadherin by Immunohistochemistry Loss of m6A results in diminished WNT signaling&signs of perturbed HF fate.(A) Left panel: representative images from E17.5 sagittal sections immunolabeled for LEF1&PCAD (scale bars: 50 µm). White solid lines denote skin surface&dashed lines denote dermal-epidermal border. Right panel: quantification of LEF1 immunofluorescence at indicated compartments (for each condition n = 3 biological replicates ×7 images per replicate, *p<0.05&***p<0.001 by unpaired two-tailed Student’s t-test). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
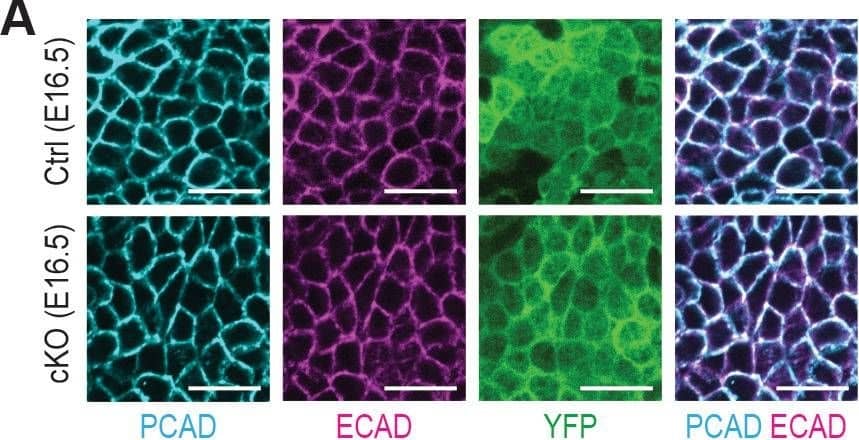
Detection of Mouse P-Cadherin by Immunohistochemistry Additional analysis of epidermal perturbations upon Mettl3 cKO.(A) Confocal images of E16.5 whole-mount back skin immunolabeled for PCAD, ECAD&YFP (scale bars: 20 µm). PCAD&ECAD expression was quantified in the basal progenitors of skin epithelium (middle line corresponds to the median; for each condition the data are from two biological replicates; ****p<0.0001 by Mann Whitney test). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
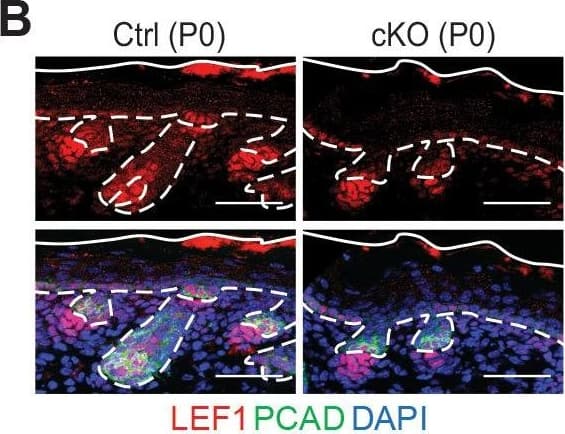
Detection of Mouse P-Cadherin by Immunohistochemistry Loss of m6A results in diminished WNT signaling&signs of perturbed HF fate. (B) Left panel: representative images from P0 sagittal sections immunolabeled for LEF1&PCAD (scale bars: 50 µm). Solid&dashed lines as in (A). Right panel: quantifications of LEF1 immunofluorescence in indicated compartments (for each condition n = 3 biological replicates ×7 images per replicate, *p<0.05&***p<0.001 by unpaired two-tailed Student’s t-test). Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
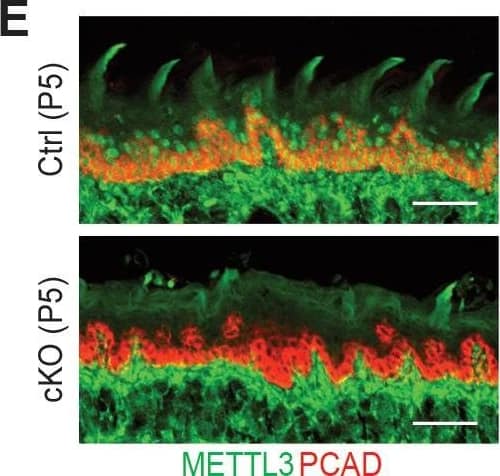
Detection of Mouse P-Cadherin by Immunohistochemistry Additional information on the Mettl3 cKO phenotypes.(A) Representative pictures of P6 control and cKO littermates (scale bars: 1 cm). (B) Quantification of body weights of control and cKO littermates (error bars: standard deviation, for each condition n = 5 biological replicates from three litters). (C) Measurements of TEWL (error bars: standard deviation, for each condition n = 4 biological replicates, for the comparison between control and cKO at each time point, p>0.05 by unpaired two-tailed Student’s t-test). (D) Representative pictures of P5 pups’ mouths demonstrating the growth of the teeth (black arrows). (E) Images of P5 tongue sagittal sections immunolabeled for METTL3 and PCAD demonstrating the tongue surface of the Mettl3 cKO animals tends to be smoother than that of the control animals, which suggests filiform papillae are not well-formed in the cKO animals (scale bars: 50 µm). (F) Hematoxylin and eosin (H and E) stained P6 back skin sagittal sections (scale bars: 100 µm).Figure 2—figure supplement 2—source data 1.Quantification of neonates' body weights in (B).Figure 2—figure supplement 2—source data 2.Quantification of TEWL in (C).Quantification of neonates' body weights in (B).Quantification of TEWL in (C). Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
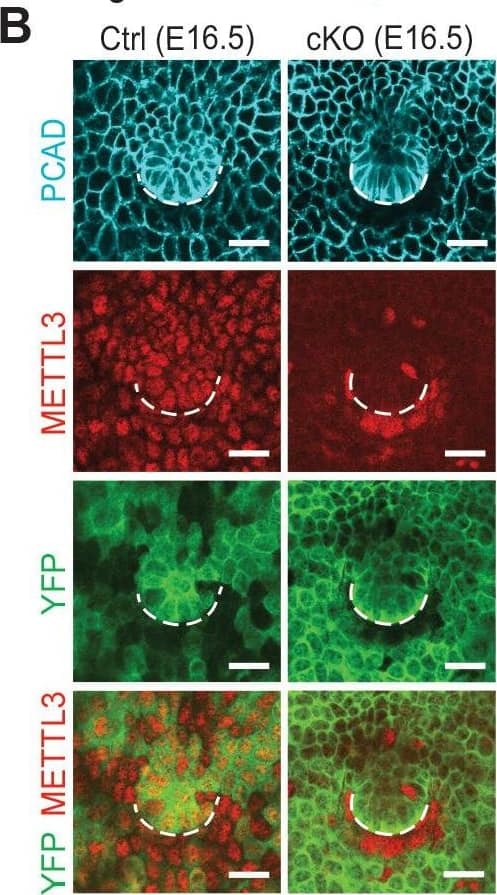
Detection of Mouse P-Cadherin by Immunohistochemistry Krt14-Cre driven conditional Mettl3 knockout mice display severe defects in HF morphogenesis. (B) Confocal images of E16.5 whole-mount back skin immunolabeled for P-cadherin (PCAD), METTL3&YFP (scale bars: 20 µm). Note that nuclear METTL3 immunofluorescence is selectively depleted from the YFP+ cells in cKO skin. White dashed lines denote the dermal-epidermal border. Image collected & cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32845239), licensed under a CC-BY license. Not internally tested by R&D Systems.
Preparation and Storage
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: P-Cadherin
Placental Cadherin (P-Cadherin or PCAD) is a member of the cadherin family of cell adhesion molecules. Cadherins are calcium-dependent transmembrane proteins, which bind to one another in a homophilic manner. On their cytoplasmic side, they associate with the three catenins, alpha, beta, and gamma (plakoglobin). This association links the cadherin protein to the cytoskeleton. Without association with the catenins, the cadherins are non-adhesive. Cadherins play a role in development, specifically in tissue formation. They may also help to maintain tissue architecture in the adult. P-Cadherin is a classical cadherin molecule. Classical cadherins consist of a large extracellular domain which contains DXD and DXNDN repeats responsible for mediating calcium-dependent adhesion, a single-pass transmembrane domain, and a short carboxy-terminal cytoplasmic domain responsible for interacting with the catenins. Constitutive P-Cadherin expression is found in the epidermis, mesothelium, corneal epithelium, and uterine decidua. Mouse P-Cadherin is an 822 amino acid (aa) protein with a 27 aa signal sequence and a 795 aa propeptide. The mature protein begins at aa 100 and has a 542 aa extracellular region, a 27 aa transmembrane region, and a 153 aa cytoplasmic region.
- Bussemakers, M.J.G. et al. (1993) Mol. Biol. Reports 17:123.
- Overduin, M. et al. (1995) Science 267:386.
- Takeichi, M. (1991) Science 251:1451.
- Nose, A. et al. (1987) EMBO J. 6:3655.
Product Datasheets
Citations for Mouse P-Cadherin Antibody
R&D Systems personnel manually curate a database that contains references using R&D Systems products. The data collected includes not only links to publications in PubMed, but also provides information about sample types, species, and experimental conditions.
26
Citations: Showing 1 - 10
Filter your results:
Filter by:
-
An RNAi screen unravels the complexities of Rho GTPase networks in skin morphogenesis
Authors: Melanie Laurin, Nicholas C Gomez, John Levorse, Ataman Sendoel, Megan Sribour, Elaine Fuchs
eLife
-
TLR2 Regulates Hair Follicle Cycle and Regeneration via BMP Signaling
Authors: Luyang Xiong, Irina Zhevlakova, Xiaoxia Z. West, Detao Gao, Rakhylia Murtazina, Anthony Horak et al.
bioRxiv
-
SIRT 7 activates quiescent hair follicle stem cells to ensure hair growth in mice
Authors: Guo Li, Xiaolong Tang, Shuping Zhang, Meiling Jin, Ming Wang, Zhili Deng et al.
The EMBO Journal
-
Role of the soluble epoxide hydrolase in the hair follicle stem cell homeostasis and hair growth
Authors: Zumer Naeem, Sven Zukunft, Stephan Günther, Stefan Liebner, Andreas Weigert, Bruce D. Hammock et al.
Pflügers Archiv - European Journal of Physiology
-
m6A RNA methylation impacts fate choices during skin morphogenesis
Authors: Linghe Xi, Thomas Carroll, Irina Matos, Ji-Dung Luo, Lisa Polak, H Amalia Pasolli et al.
eLife
-
Correction of aberrant growth preserves tissue homeostasis
Authors: Samara Brown, Cristiana M. Pineda, Tianchi Xin, Jonathan Boucher, Kathleen C. Suozzi, Sangbum Park et al.
Nature
-
Decomposing a deterministic path to mesenchymal niche formation by two intersecting morphogen gradients
Authors: Qu R, Gupta K, Dong D et al.
Developmental cell
-
Progenitors oppositely polarize WNT activators and inhibitors to orchestrate tissue development
Authors: Irina Matos, Amma Asare, John Levorse, Tamara Ouspenskaia, June de la Cruz-Racelis, Laura-Nadine Schuhmacher et al.
eLife
-
Identification of hair shaft progenitors that create a niche for hair pigmentation
Authors: Chung-Ping Liao, Reid C. Booker, Sean J. Morrison, Lu Q. Le
Genes & Development
-
Signalling by senescent melanocytes hyperactivates hair growth
Authors: Wang, X;Ramos, R;Phan, AQ;Yamaga, K;Flesher, JL;Jiang, S;Oh, JW;Jin, S;Jahid, S;Kuan, CH;Nguyen, TK;Liang, HY;Shettigar, NU;Hou, R;Tran, KH;Nguyen, A;Vu, KN;Phung, JL;Ingal, JP;Levitt, KM;Cao, X;Liu, Y;Deng, Z;Taguchi, N;Scarfone, VM;Wang, G;Paolilli, KN;Wang, X;Guerrero-Juarez, CF;Davis, RT;Greenberg, EN;Ruiz-Vega, R;Vasudeva, P;Murad, R;Widyastuti, LHP;Lee, HL;McElwee, KJ;Gadeau, AP;Lawson, DA;Andersen, B;Mortazavi, A;Yu, Z;Nie, Q;Kunisada, T;Karin, M;Tuckermann, J;Esko, JD;Ganesan, AK;Li, J;Plikus, MV;
Nature
Species: Transgenic Mouse
Sample Types: Whole Tissue
Applications: Immunohistochemistry -
Stem cells tightly regulate dead cell clearance to maintain tissue fitness
Authors: Stewart, KS;Gonzales, KA;Yuan, S;Tierney, MT;Bonny, AR;Yang, Y;Infarinato, NR;Cowley, CJ;Levorse, JM;Pasolli, HA;Ghosh, S;Rothlin, CV;Fuchs, E;
bioRxiv : the preprint server for biology
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Transcriptomic Changes Predict Metabolic Alterations in LC3 Associated Phagocytosis in Aged Mice
Authors: A Dhingra, JW Tobias, NJ Philp, K Boesze-Bat
International Journal of Molecular Sciences, 2023-04-04;24(7):.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Sox2 in the dermal papilla regulates hair follicle pigmentation
Authors: KJ Ng, J Lim, YN Tan, D Quek, Z Lim, N Pantelirei, C Clavel
Oncogene, 2022-07-19;40(3):111100.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Corticosterone inhibits GAS6 to govern hair follicle stem-cell quiescence
Authors: S Choi, B Zhang, S Ma, M Gonzalez-C, D Stein, X Jin, ST Kim, YL Kang, A Besnard, A Rezza, L Grisanti, JD Buenrostro, M Rendl, M Nahrendorf, A Sahay, YC Hsu
Nature, 2021-03-31;0(0):.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Distinct Regulatory Programs Control the Latent Regenerative Potential of Dermal Fibroblasts during Wound Healing
Authors: S Abbasi, S Sinha, E Labit, NL Rosin, G Yoon, W Rahmani, A Jaffer, N Sharma, A Hagner, P Shah, R Arora, J Yoon, A Islam, A Uchida, CK Chang, JA Stratton, RW Scott, FMV Rossi, TM Underhill, J Biernaskie
Cell Stem Cell, 2020-08-04;27(3):396-412.e6.
Species: Mouse, Transgenic Mouse
Sample Types: Whole Cells, Whole Tissue
Applications: ICC, IHC -
miR-29a/b1 Inhibits Hair Follicle Stem Cell Lineage Progression by Spatiotemporally Suppressing WNT and BMP Signaling
Authors: M Ge, C Liu, L Li, M Lan, Y Yu, L Gu, Y Su, K Zhang, Y Zhang, T Wang, C Liu, F Liu, M Li, L Xiong, K Wang, T He, Y Dai, Y Zhao, N Li, Z Yu, Q Meng
Cell Rep, 2019-11-19;29(8):2489-2504.e4.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Functionally Distinctive Ptch Receptors Establish Multimodal Hedgehog Signaling in the Tooth Epithelial Stem Cell Niche
Authors: M Binder, P Chmielarz, PJ Mckinnon, LC Biggs, I Thesleff, A Balic
Stem Cells, 2019-06-10;0(0):.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Cadherins in the retinal pigment epithelium (RPE) revisited: P-cadherin is the highly dominant cadherin expressed in human and mouse RPE in vivo
Authors: X Yang, JY Chung, U Rai, N Esumi
PLoS ONE, 2018-01-16;13(1):e0191279.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Stem cell plasticity enables hair regeneration following Lgr5(+) cell loss
Authors: JD Hoeck, B Biehs, AV Kurtova, NM Kljavin, F de Sousa E, B Alicke, H Koeppen, Z Modrusan, R Piskol, FJ de Sauvage
Nat. Cell Biol., 2017-05-29;19(6):666-676.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Stem Cell Lineage Infidelity Drives Wound Repair and Cancer
Authors: Y Ge, NC Gomez, RC Adam, M Nikolova, H Yang, A Verma, CP Lu, L Polak, S Yuan, O Elemento, E Fuchs
Cell, 2017-04-20;0(0):.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC -
Role of cell and matrix-bound VEGF isoforms in lens development.
Authors: Saint-Geniez M, Kurihara T, D'Amore PA
Invest. Ophthalmol. Vis. Sci., 2008-08-29;50(1):311-21.
Species: Mouse
Sample Types: Whole Tissue
Applications: IHC-Fr -
Single-Cell Analysis Reveals a Hair Follicle Dermal Niche Molecular Differentiation Trajectory that Begins Prior to Morphogenesis
Authors: Gupta K, Levinsohn J, Linderman G et al.
Dev. Cell
-
E‐cadherin mediates apical membrane initiation site localisation during de novo polarisation of epithelial cavities
Authors: Xuan Liang, Antonia Weberling, Chun Yuan Hii, Magdalena Zernicka‐Goetz, Clare E Buckley
The EMBO Journal
-
Keratin-mediated hair growth and its underlying biological mechanism
Authors: Seong Yeong An, Hyo-Sung Kim, So Yeon Kim, Se Young Van, Han Jun Kim, Jae-Hyung Lee et al.
Communications Biology
-
Loss of lamin B1 is a biomarker to quantify cellular senescence in photoaged skin
Authors: Audrey Shimei Wang, Peh Fern Ong, Alexandre Chojnowski, Carlos Clavel, Oliver Dreesen
Scientific Reports
-
Molar Bud-to-Cap Transition Is Proliferation Independent
Authors: S. Yamada, R. Lav, J. Li, A.S. Tucker, J.B.A. Green
Journal of Dental Research
FAQs
No product specific FAQs exist for this product, however you may
View all Antibody FAQsReviews for Mouse P-Cadherin Antibody
There are currently no reviews for this product. Be the first to review Mouse P-Cadherin Antibody and earn rewards!
Have you used Mouse P-Cadherin Antibody?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image
